ByteDance Releases Protenix-v1: A New Open Source Model That Achieves AF3-Level Performance in Biomolecular Structure Prediction

How close can an open model get to AlphaFold3 level accuracy if it matches the training data, model scale and decision budget? ByteDance presented Protenix-v1perfect Reproduction of AlphaFold3 (AF3). for the prediction of biomolecular structure, extracted with code and model parameters under Apache 2.0. The model is directed AF3 level performance for all protein, DNA, RNA and ligand structures while keeping the entire stack open and scalable for research and production.
The main release is also shipped with PXMeter v1.0.0an assessment toolkit and its data set transparent benchmarking on more than 6k complexes with time segmentation and domain-specific subsets.
What is Protenix-v1?
Protenix is defined as ‘Protenix: Protein + X‘, the basic model of high accuracy of biomolecular structure. It is predictable 3D atomic structures of complexes can include:
- Proteins
- Nucleic acids (DNA and RNA)
- Ligands are small molecules
The research team describes Protenix as complete reproduction of AF3. It reuses the AF3-style distribution structure for all atomic types and exposes it to a trainable PyTorch codebase.
The project is released as a full stack:
- Training code and description
- Pre-trained model weights
- MSA data and pipelines
- Supported browser Protenix web server interactive use
AF3 standard performance under similar constraints
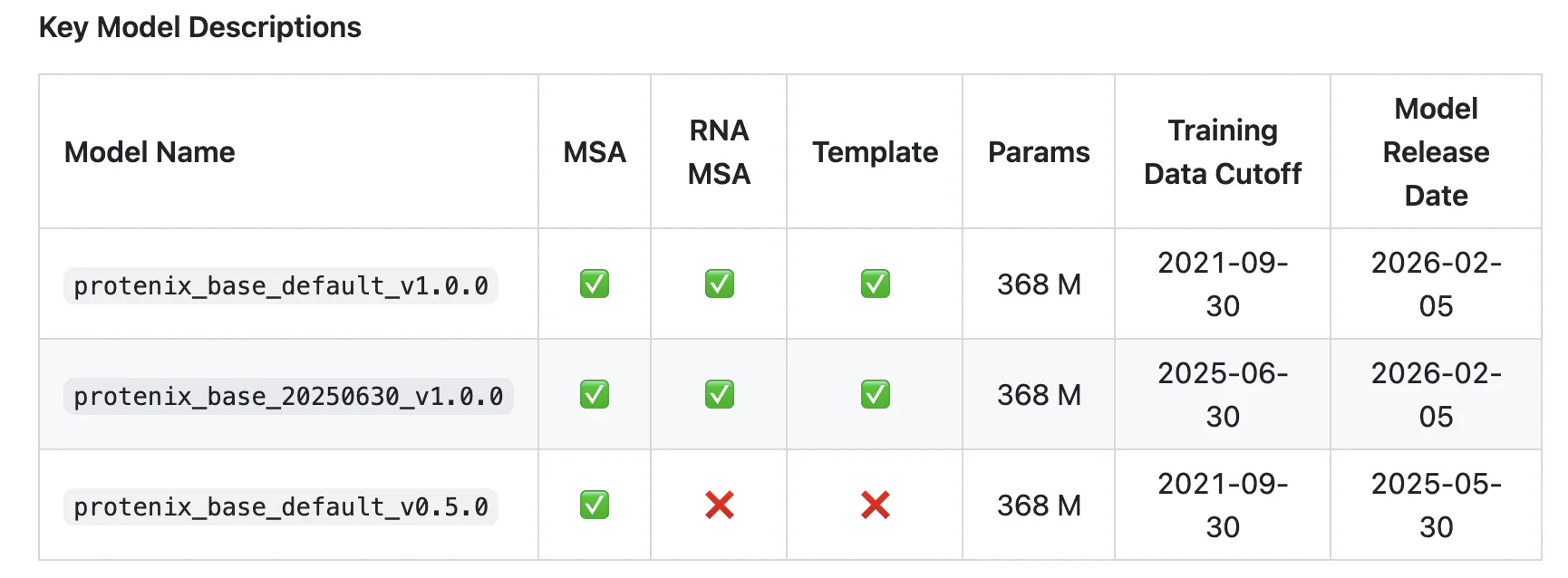
According to the research team Protenix-v1 (protenix_base_default_v1.0.0) is something ‘the first fully open source model that outperforms AlphaFold3 on all different measurement sets while adhering to the same training data cutoff, model scale, and targeting budget as AlphaFold3.‘
The main obstacles are:
- Termination of training data: 2021-09-30, aligned with AF3’s PDB cutoff.
- Model scale: Protenix-v1 itself has 368M parameters; AF3 scale is modeled but not disclosed.
- Fixed budget: the comparison uses the same sample budgets and time constraints.
For challenging objectives such as antigen-antibody complexesthe increase of number of sample candidates from several to hundreds of fruits a log-linear constant improvement in accuracy. This provides a clear and written explanation Inference-time scaling behavior instead of one fixed workplace.
PXMeter v1.0.0: Testing 6k+ builds
To support these claims, the research team released PXMeter v1.0.0an open source toolkit for benchmarks for predicting a reproducible structure.
PXMeter offers:
- A a hand-picked benchmark data setwith non-biological artifacts and problematic entries removed
- Time segmentation and domain-specific subsets (eg, antibody-antigen, protein-RNA, ligand complexes)
- A a unified assessment framework which includes metrics such as complex LDDT and DockQ for all models
Related PXMeter research paper, ‘Revisiting benchmarks for structural prediction with PXMeter,‘ it checks Protenix, AlphaFold3, Boltz-1 and Chai-1 on the same selected tasks, and shows how different dataset designs affect the model’s quality and perceived performance.
How Protenix fits into the broader stack?
Protenix is part of a small ecosystem of related projects:
- PXDesign: a binder design suite built on the Protenix base model. It reports 20–73% test hit rates again 2–6× higher success there are methods like AlphaProteo and RFdiffusion, and it is accessible through Protenix Server.
- Protenix-Dock:a the classical protein-ligand docking framework which uses scoring functions instead of deep nets, tuned by strong docking functions.
- Protenix-Mini and such follow-up work Protenix-Mini+: a lightweight alternative that reduces the cost of inference using compression of structures and sampling of a few step distributions, while maintaining an accuracy within a few percent of the full model in standard benchmarks.
Together, these components cover structural forecasting, shipping, and design, and share interfaces and formats, making it easy to integrate into pipelines.
Key Takeaways
- AF3-class, fully open model: Protenix-v1 is an open-source AF3-style all-atom biomolecular structure predictor with weights under Apache 2.0, targeting proteins, DNA, RNA and ligands.
- AF3 tight alignment for good comparison: Protenix-v1 is similar to AlphaFold3 in important axes: training data cut-off (2021-09-30), model scale class and thin imaging budget, which allows proper AF3 level performance claims.
- Transparent measurement with PXMeter v1.0.0: PXMeter offers a curated benchmark of over 6k+ structures with time-resolved and domain-specific subsets and integrated metrics (eg, complex LDDT, DockQ) for retesting.
- A proven treatment for measuring inference time: Protenix-v1 shows log-linear accuracy gains as the number of candidate samples increases, providing a documented delay—trading accuracy over a single fixed operating point.
Check it out Repo and Try it here. Also, feel free to follow us Twitter and don’t forget to join our 100k+ ML SubReddit and Subscribe to Our newspaper. Wait! are you on telegram? now you can join us on telegram too.