Google DeepMind Unveils AlphaGenome: An Integrated Sequence-to-Function Model Using Hybrid Transformers and U-Nets to Extract the Human Genome
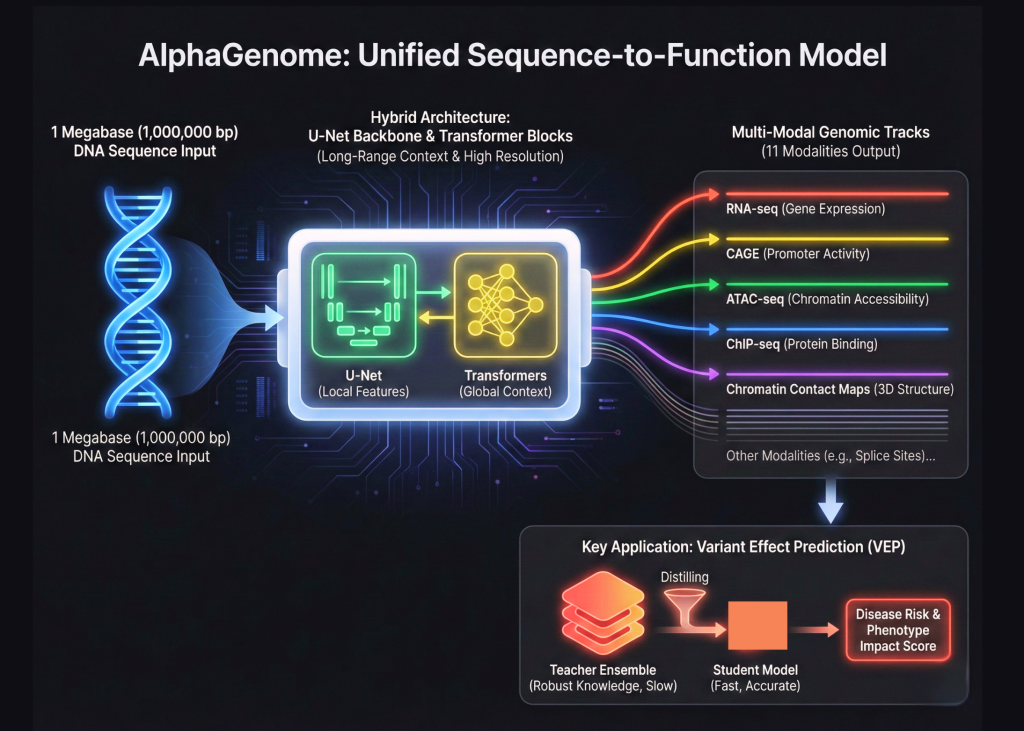
Google DeepMind is expanding its biological toolkit beyond the world of protein folding. After the success of AlphaFold, a team of Google researchers launched AlphaGenome. This is a unified deep learning model designed for functional sequencing of genomics. This represents a major change in the way we model the human genome. AlphaGenome does not treat DNA as plain text. Instead, it processes 1,000,000 base windows of raw DNA to predict the cell’s functional state.
Bridging the Scale Gap with Hybrid Architectures
The complexity of the human genome comes from its scale. Many existing models struggle to see the big picture while tracking the fine details. AlphaGenome solves this by using a hybrid architecture. Includes U-Net backbone and Transformer blocks. This allows the model to capture long-range interactions across 1 Megabase of sequence while maintaining base pair resolution. This is like building a system that can read a thousand page book and still remember the exact location of a single comma.
Mapping Sequences to Biologically Active Species
AlphaGenome is a functional model sequencer. This means that its main goal is to map DNA sequences directly to biological functions. These activities are measured by genomic tracks. The research team trained AlphaGenome to predict 11 different genomic pathways. These methods include RNA-seq, CAGE, and ATAC-seq. They also include ChIP-seq of various transcription factors and chromatin interaction maps. By predicting all these tracks simultaneously, the model gains a complete understanding of how DNA controls the cell.
The Power of Multitasking Learning in Genomics
AlphaGenome’s technological breakthrough lies in its ability to handle 11 different types of data simultaneously. In the past, researchers often developed separate models for each task. AlphaGenome uses a multi-task learning approach. This helps the model learn shared characteristics across different biological processes. If a model understands how a protein binds to DNA, it can better predict how that DNA will be expressed as RNA. This integrated approach reduces the need for multiple specialized models.
Improving Differential Effect Prediction by Distillation
One of AlphaGenome’s most important programs is Variant Effect Prediction, or VEP. This process determines how a single mutation in DNA affects the body. Mutations in genes can lead to diseases such as cancer or heart disease. AlphaGenome excels in this by using a specific training method called Teacher Student distillation. The research team first developed a set of ‘whole’ teacher models. These teachers are trained on large amounts of genomic data. After that, they break down that information into a model for one student.
Critical Information for Precision Medicine
This distillation process makes the model faster and more robust. This is a common way to compress information. However, applying it to genomics at this scale is a new milestone. The learner model learns to replicate the high-quality predictions of the teacher’s ensemble. This allows it to identify dangerous mutations with high accuracy. The model can also predict how mutations in distant genes can affect a DNA strand.
High-Performance Computing with JAX and TPUs
The architecture is implemented using JAX. JAX is a highly functional computer code library. It is often used for advanced machine learning at Google. Using JAX allows AlphaGenome to run efficiently on Tensor Processing Units, or TPUs. The research team used sequence parallelism to handle large input 1 Megabase windows. This ensures that memory requirements do not increase as the sequence length increases. This shows the importance of choosing the right framework for big biological data.
Transfer Learning About Data Cell Types
AlphaGenome also faces the challenge of missing data on certain cell types. Because it is a basic model, it can be optimized for specific tasks. The model learns general biological rules from large public datasets. These rules may be applied to rare diseases or certain tissues where data are difficult to obtain. This transfer learning ability is one of the reasons why AlphaGenome is so versatile. It can predict how a gene will behave in a brain cell even if it was trained primarily on liver cell data.
Towards a New Era of Personalized Care
In the future, AlphaGenome could lead to a new era of personalized medicine. Doctors can use the model to scan a patient’s entire genome in 1,000,000 base pair chunks. They can pinpoint which variants may cause health problems. This will allow treatments tailored to a specific individual’s genetic code. AlphaGenome brings us closer to this reality by providing a clear and accurate map of the working genome.
Setting the Standard for Biological AI
AlphaGenome also marks the AI revolution in genomics. It proves that we can model very complex biological systems using the same principles used in modern AI. By combining the architecture of U-Net and Transformers and using student immersion for teachers, the Google DeepMind team has set a new standard.
Key Takeaways
- Hybrid Sequence Architecture: AlphaGenome uses a special hybrid design that includes a I-Net spine with The converter blocks. This allows the model to process large windows of 1,000,000 base pairs while maintaining the high resolution required to identify a single mutation.
- Functional Forecasting for Many Situations: The model is trained to predict 11 different genomic approaches simultaneously, including RNA-seq, CAGE, and ATAC-seq. By studying these various biological tracks together, the program gains a comprehensive understanding of how DNA controls cellular activity in all different tissues.
- Teacher-student Distillation: To achieve industry-leading precision Varian Effect Prediction (VEP)researchers used a digestion method. They transferred knowledge from a collection of high-performing ‘teacher’ models to a single, efficient ‘learner’ model that is faster and more robust at identifying disease-causing mutations.
- Designed for High Performance Computing: The frame is used in JAX and prepared TPUs. By using sequencing, AlphaGenome can handle the computational load of megabase-scale DNA sequence analysis without exceeding memory limits, making it a powerful tool for large-scale research.
Check it out Paper again Repo. Also, feel free to follow us Twitter and don’t forget to join our 100k+ ML SubReddit and Subscribe to Our newspaper. Wait! are you on telegram? now you can join us on telegram too.



